Research Description
Research Description UPDATE COMING SOON!!
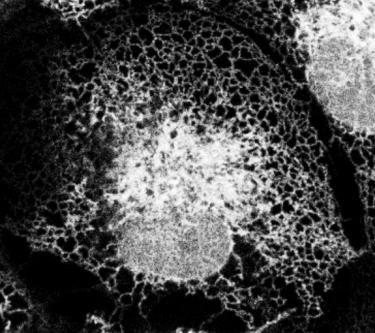
We study the architecture and dynamics of the endoplasmic reticulum (ER), the largest organelle in cells that has essential functions in lipid and protein synthesis. The ER is comprised of multiple structural and functional domains, the most prominent of which is the nuclear envelope. While much is known about the protein machinery that generates the shape of the ER, less is known about the nanoscale membrane structures and lipid chemistries that give rise to its domain organization.
To address these open areas we take a multi-disciplinary approach that combines our expertise in quantitative high resolution fluorescent live imaging and genome engineering (both in vertebrate cells and C. elegans) with the expertise of our collaborators in super resolution microscopy and biophysical approaches.
______________________________________________________________________________________________
It is known that interactions with the cytoskeleton and membrane shaping proteins are essential to shape the ER and nuclear envelope. A less explored but emerging area is an understanding of how the chemistry and amount of bilayer lipids that make up the membranes themselves are controlled to establish and maintain the unique domain organization and nanoscale membrane structures of the ER/nuclear envelope. This has become an exciting area to explore as our understanding of the membrane architecture of the ER continues to expand with advances in light microscopy. This is highlighted by our collaborative work with the Bewersdorf lab that revealed dynamic nano-scale size holes in ER sheets using a type of super resolution microscopy called STED (STimulated Emission Depletion) (Schroeder et al. 2019).
We have contributed to this area by showing that de novo lipid synthesis is regulated during the cell cycle to restrict ER sizeand maintain ER architecture as cells grow and prepare for cell division. ER size impacts its reorganization and in turn the transmission of the genome by controlling the biophysical properties of the mitotic cytoplasm (Merta & Carrasquillo-Rodriguez et al. 2021). The production of membranes by lipid synthesis also coordinates with the sealing of nuclear membranes during nuclear formation and controls the rate of nuclear growth (Penfield et al. 2018 & 2020, Mauro et al. 2022). Furthermore, we discovered that bilayer lipid chemistries of the ER and nuclear envelope contribute directly to regulating the proteins that associate with their membranes and execute their functions (Lee et al. 2022).
Together, our work has revealed that the abundance and chemistry of bilayer lipids that constitute the membranes of the ER are highly regulated to give rise to the domain organization of the ER. The nuclear envelope domain protects the genome and ER reorganization is critical to transmission of the genome and so our work links dysregulation of lipid metabolism to genomic stability, hallmarks of cancer cells.
______________________________________________________________________________________________
Current projects:
1. Understanding the distribution, content, and regulation of lipids and lipid metabolizing enzymes throughout the ER and how it relates to ER/nuclear envelope organization is a major goal of the lab. In addition to our live imaging approaches, we use protein modeling, structure-function analyses, endogenous tagging by CRISPR-Cas9 and different cell types (in both in C elegans and mammalian cells) to gain a deep mechanistic understanding of the targeting and regulation of enzymes involved in lipid metabolism at the ER/nuclear envelope.
2. The nuclear envelope is arguably the most distinct domain of the ER. It has a unique protein composition to execute its distinct functions. Resident nuclear envelope proteins are regulated by the biophysical properties of nuclear membranes, which is determined largely by its bilayer lipid composition. Isolating the nuclear envelope from the rest of the ER for biochemical analysis of its lipid make-up is challenging because of the direct continuity of their membranes. We are addressing this completely open area by developing and employing a variety of genetically encoded and chemical tools to locally detect and manipulate certain types of lipids and determine the functional consequences. We are also screening for lipid sensing domains in ER/nuclear envelope proteins to further probe the bilayer lipid distributions and compositions important to their regulation and functions.
3. The domain organization and architecture of the ER and nuclear envelope is completely dismantled during cell division. We study the regulation and functional effects of mitotic ER reorganization, with a specific focus on nuclear envelope breakdown and reformation. We are currently studying how membranes detach from condensing chromosomes upon entry into mitosis and how membranes enwrap chromosomes to form a nucleus at the end of mitosis. Within this theme, we have projects on the process of membrane remodeling to form a sealed nuclear compartment in both mammalian cells and C. elegans.